Nervous System collaborates on new 3D-printed organ research published on cover of Science
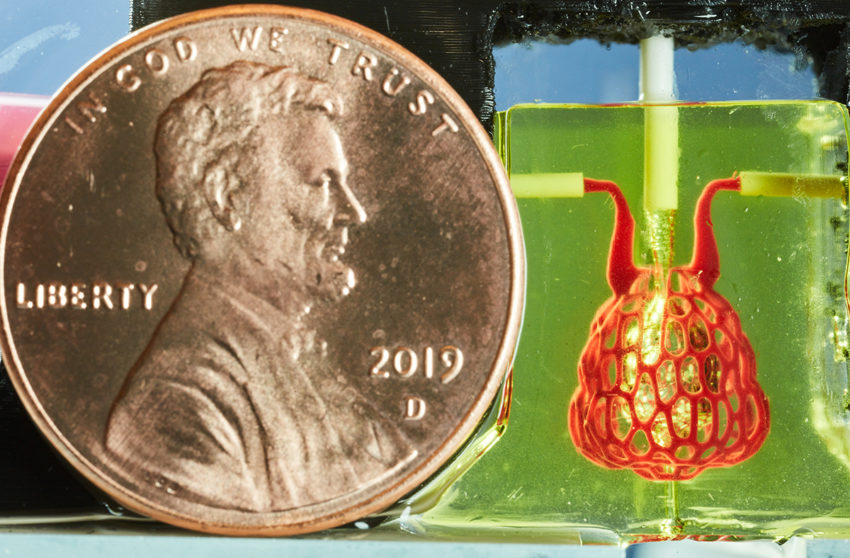

We founded Nervous System in 2007 with the goal of adapting algorithms and strategies from nature into new ways to design products; but we never imagined we’d have the opportunity to apply that back to designing living things. For the past 3 years we’ve collaborated with Jordan Miller, a bioengineer at Rice University, on creating vascular networks for 3D-printed organs. Today our work appears on the cover of Science Magazine where it is published in an interdisciplinary article which we co-authored. The work was led by Jordan Miller at Rice and Kelly Stevens at the University of Washington and included 13 additional collaborators from Rice, University of Washington, Duke University, and Rowan University.
It all began with an email from Jordan Miller in February 2016. Jordan was familiar with us from the open source models we had posted on Thingiverse, and in his words “totally captivated” by the series of branching sculptures we created for our growing objects exhibit. He proposed that our skills could be put to an epic task: “perhaps we could work together to make open-source software that the world could use to design synthetic living tissues and organ replacements for human patients.” The idea of taking our generative systems which are inspired by nature and using them to actually make living things was a dream come true.

DOI: 10.1126/science.aav9750 . Reprinted with permission from AAAS.
While there are many labs creating artificial tissues, the problem is keeping them alive. Organs like the liver are composed of more than 100 billion cells which need support from intricate vascular networks that provide nutrients and oxygen while removing waste. Making things even more complicated, many organs contain multiple interpenetrating networks of fluids; in the lungs we have hierarchically branched airways and ensheathing blood vessel networks. Our goal is to create software which enables scientists to design customized multi-vascular structures for 3D-printed organs. Nervous System collaborated with the Miller Lab to design and generate these complex multivascular networks and materialize them in soft hydrogels for the first time.
A new 3D-printing method for cells
Our role is just a small part of a large team led by Jordan Miller at Rice University. The Miller Lab is developing the bioengineering, 3D-printing technology, cell culturing, and analysis tools that make these designs possible to realize. Particularly, they developed a new 3D-printing workflow compatible with live cells called SLATE (stereolithography apparatus for tissue engineering). The SLATE printer can embed live cells into soft gels containing very small, intricate blood vessels down to 300 microns in diameter. Hydrogels printed in only minutes by SLATE can function as lung-like networks with entangled air / blood networks. We got to see the process firsthand during trips to Jordan’s lab in Houston and they are truly amazing.
This new printing method allows for new and dramatic architectural freedoms in the living tissues we can design and fabricate. Bioengineers don’t have tools at their disposal to generate these complex architectures that are crucially needed to keep living tissues alive. That’s where we came in.
Designing for living tissues
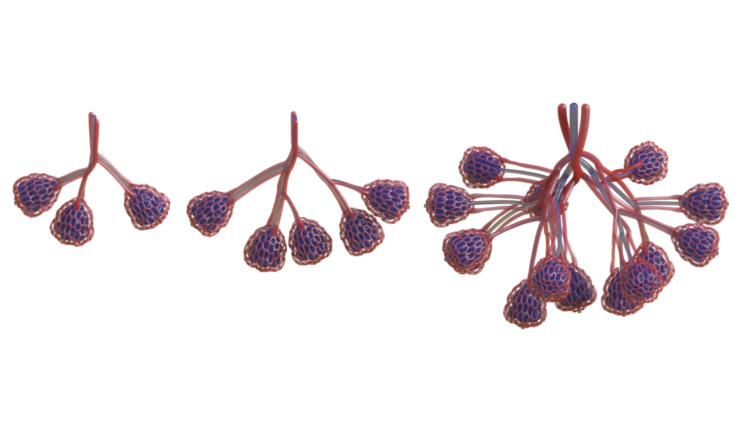
Nervous System collaborated with the Miller lab to design vascular networks and developed custom software to generate lung-mimetic designs for 3D-printing engineered living tissues. This software makes it possible to generate entangled vessel networks within any user-defined volume and connect these networks to inlets and outlets so air and blood can flow through the intertwined networks.
We grow the branching form of the airway algorithmically. It reaches out into a volume defined by the designer, colonizing it evenly with air conduits while connecting back to the initial growth point like a tree. Air is pumped into the network and it pools at the bulbous air sacs which crown each tip of the network. These sacs are rhythmically inflated and deflated by breathing action, so called tidal ventilation because the air flow in human lungs is reminiscent of the flows of the ocean tides.
Next we grow dual networks of blood vessels that entwine around the airway. One to bring deoxygenated blood in, the other to carry oxygen-loaded blood away. The two networks join at the tips of the airway in a fine mesh of blood vessels which ensheathes the bulbous air sacs. These vessels are only 300 microns wide!
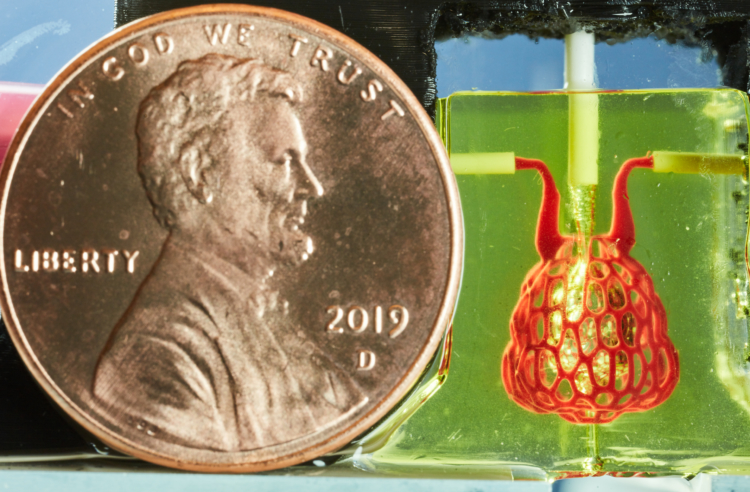
So far we’ve printed and tested the alveolar subunits of this system but we have designs for printing much larger networks of them that could be tested for their efficiency and potential as lung replacements. What is printable lags behind what we can design at this point. It took us three days to generate the lung-mimicking architecture but it took about a year until it was possible to fabricate it.
To build this software, we cobbled together some portions of previous generative systems we’ve written and created new tools as well. The growth of the airway builds upon our Hyphae 3D software. The directional cellular vessels on the air sacs are related to our Corollaria software. However the way we grew the intertwining vessel networks that run parallel to the airway is something new which we describe below.
Designing the Alveoli-like unit
The architecture we designed for the blood vessel network was inspired by the cellular capillaries which surround alveoli. We designed them using an anisotropic, centroidally optimized voronoi diagram offset from the surface of the air sac. The anisotropy elongates the cells along the direction from the inlet to the outlet to encourage fluid flow around the air sac instead of taking a shortcut right to the blood outlet. The centroidal optimization produces a network which is ideal for distributing resources.
The Miller Lab fabricated and tested the architectures we generated showing that they can withstand more than 10,000 ventilation cycles while being perfused with human red blood cells. Study of the printed gels shows that the architecture we designed promotes red blood cells mixing and bidirectional flow which is hypothesized to occur in the human lung.
Designing the entangled vasculature
The entwining blood vessel network is generated parallel to the airway. We create an offset network by building a frame field along the airway. The frame field is defined at nodal connections as normal to the plane defined by the 3 points of that node. Everywhere else the frame field is interpolated by parallel transporting the nodal frame field to its children along the network. The entwining network is produced by offsetting the airway in the direction of the frame field a distance proportional to the thickness of the network.
As artists and designers, our work often appears in galleries or shops but today it appears on the cover of one of the most well-respected scientific journals, Science, a publication of the American Association for the Advancement of Science (AAAS). We feel very lucky to have had the chance to participate in this ground breaking research and are excited to keep working with Jordan Miller and his team on this project going forward.
The paper is available here: Multivascular networks and functional intravascular topologies within biocompatible hydrogels
Project credits
- Rice University: Bagrat Grigoryan, Samantha Paulsen, Daniel Sazer, Alexander Zaita, Paul Greenfield, Nicholas Calafat, Anderson Ta, and Jordan Miller;
- University of Washington: Daniel Corbett, Chelsea Fortin, Fredrik Johansson and Kelly Stevens;
- Duke University: John Gounley and Amanda Randles
- Nervous System: Jessica Rosenkrantz and Jesse Louis-Rosenberg
- Rowan University: Peter Galie
The work was supported by the Robert J. Kleberg Jr. and Helen C. Kleberg Foundation, the John H. Tietze Foundation, the National Science Foundation (1728239, 1450681 and 1250104), the National Institutes of Health (F31HL134295, DP2HL137188, T32EB001650, T32GM095421 and DP5OD019876) and the Gulf Coast Consortia.




Bronwyn Johnson
So completely overjoyed to read about this. I love it when art and science combine to realise a greater goal.
Stefan Guggenberger
Nice article
Richard R.
The new frontier, small and beautiful, and life-saving.